
JC Jang
Affiliate Professor (Horticulture and Crop Sciences)
013 Rightmire Hall
1060 Carmack Rd
Columbus, OH
43210
Areas of Expertise
- Sugar and hormone signal transduction in plants; Function an
Our lab is interested in understanding the molecular mechanisms underlying sugar sensing and signal transduction in plants [1, 2]. In the early work, we showed that hexokinase is involved in sugar signal transduction [3]. We then showed that both hexokinase-dependent and hexokinase-independent sugar signal transduction mechanisms exist in plants [4]. We also helped to demonstrate that plant sugar response can be modulated by stress hormone, hence forging the concept of sugar-stress hormone crosstalk [5]. Later we found that sugar induced stress hormone accumulation is partly responsible for sugar-stress hormone crosstalk [6]. Based on the results of recent transcriptome analyses, we proposed a complex sugar signaling network mediated by multiple signal transduction pathways [7].
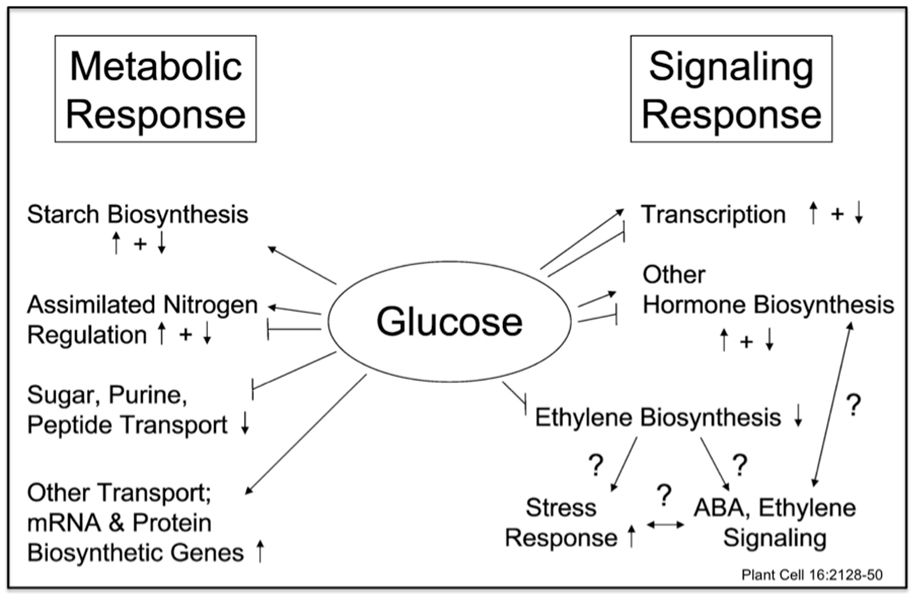
To further decipher this network, we began to systematically characterize sugar responsive transcription factors for their roles in various sugar signaling mechanisms. We showed that sugar responsive transcription factor AtbZIP1 could form a complex bZIP protein dimerization network that differentially regulates downstream sugar responsive genes [8]. With recent new findings on another group of sugar responsive transcription factors, our current research is focused on:
-
The functional analysis of Arabidopsis thaliana Tandem Zinc Finger (TZF) genes.
-
Revealing the role of AtTZFs in plant P-body and stress granule function and assembly.
Plant Tandem Zinc Finger (TZF) genes
Sugar sensing and signal transduction is pivotal in coordinating growth and stress responses in plants. Our transcriptome analysis has uncovered 185 sugar responsive nuclear transcription factors in Arabidopsis thaliana [7]. Unexpectedly, one of the transcription factors, Tandem Zinc Finger1 (AtTZF1), can traffic between nucleus and cytoplasmic foci. These foci are characteristic of recently discovered mammalian Processing-bodies (P-bodies or PBs) and Stress Granules (SGs). PBs and SGs are cytoplasmic messenger ribonucleoprotein (mRNP) complexes, playing important roles in post-transcriptional regulation and epigenetic modulation of gene expression. While PBs are enriched with translation-repressed mRNAs and mRNA silencing and degradation machineries, SGs are super-complexes of stalled pre-initiation complexes and translation regulators. Notably, AtTZF1-associated PBs and SGs are assembled dynamically in specific plant tissues under particular stresses. They are preferentially formed in stomata and stem cells, and can be induced by stress hormone methyl jasmonate [9]. Reverse genetic analyses indicate that AtTZF1 acts as a positive regulator of sugar and ABA (stress) response, and a negative regulator of GA (growth) response. Plants over-expressing AtTZF1 are compact, late flowering, and superior in cold- and drought-tolerance (Figure 1). In addition, ABA-, Salicylic Acid- (SA-), and sugar-responsive genes are up regulated, whereas GA-responsive genes are down regulated in AtTZF1 OX plants [10].
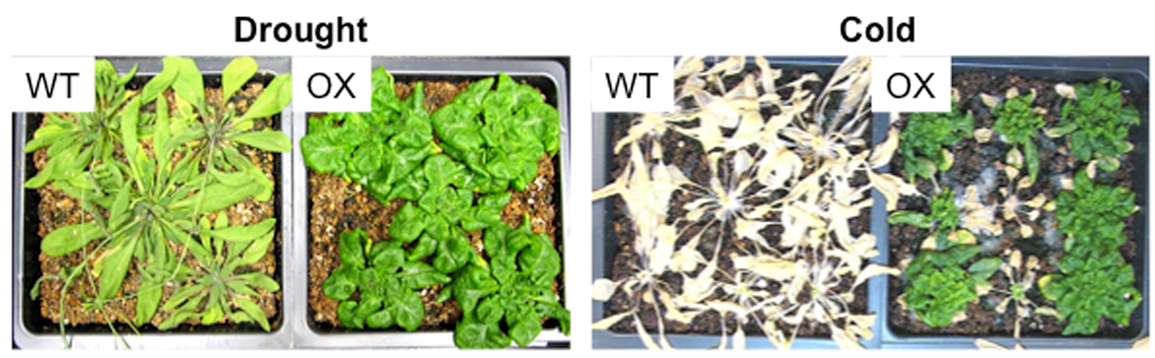
Figure 1. AtTZF1 can enhance plant stress responses. (Left Panel) Compared to the WT, AtTZF1 overexpression plants (OX) were drought tolerant, as evidenced by high survival rates under a prolonged dehydration condition. (Right Panel) Compared to the WT, OX plants were more tolerant to freezing temperatures (-9 to -11°C) under non-acclimated conditions.
In Arabidopsis, all 11 TZF subfamily IX genes (AtTZFs) contain a unique TZF motif variant that is distinct from the mammalian TZF motif. A mammalian TZF- called TTP, can induce mRNA degradation by binding to the AU-Rich Elements (AREs) at the 3’UTR of many important regulators, including Tumor Necrosis Factor a (TNF-a). Under stress conditions, TTP is concentrated in SGs and PBs, where targeted mRNAs can be stored or degraded. Although AtTZF1 can bind both DNA and RNA in vitro, it does not bind to the consensus AREs, due to the structural differences in TZF motif [9]. However, additional analysis indicates that AtTZF1 may still bind RNA elements other than consensus AREs [11]. On the basis of these results, we hypothesize that AtTZF1 may act as a transcriptional regulator in the nucleus and a RNA regulator in PBs and SGs. Work is in progress to reveal: 1) The AtTZF1 nucleic acid binding sites, 2) Mechanisms that regulate AtTZF1 intracellular trafficking, 3) Role of AtTZF1 in PB and SG assembly and function, 4) AtTZF1 associated mRNP complex.
Plant P-bodies (PBs) and stress granules (SGs)
In eukaryotic cells, gene expression is determined not only by transcription, but also by post-transcriptional events that control RNA processing, transport, stability, localization, and ultimately translation. Traditionally, it is thought that mRNAs are translated or degraded in the cytoplasm. However, when mRNAs fail to pass quality controls or translation is repressed by stress or miRNA, messenger ribonucleoprotein complexes (mRNPs) are temporarily stored in cytoplasmic foci known as Processing-bodies (P-bodies, PBs) or Stress Granules (SGs). Depending on the cellular condition, mRNAs in PBs or SGs can either re-enter the normal translation cycle or exit to the mRNA decay pathways for degradation. Hence PBs and SGs play pivotal roles in post-transcriptional regulation, as well as epigenetic modulation of gene expression [12, 13]. While PBs and SGs are distinct, they are compositionally and functionally linked. They work in concert as capacitors for mRNA metabolism under stress conditions. SGs are induced by many stresses such as heat, oxidative stress, UV irradiation, hypoxia, viral infection, and glucose starvation. PBs are often found in active growing cells and also induced by many stresses in mammalian cells. Interestingly, overexpression of a mammalian TZF, TTP, not only induces the assembly of PBs and SGs, but also promotes their physical interaction [14].
Although PBs and SGs have been studied intensively in mammals and yeast, it was unclear until recently if PBs and SGs exist in plants. In 2006-7, plant decapping complex proteins (DCP1, DCP2, and VCS/Hedls) were found in PB-like cytoplasmic foci by several research groups [15-17]. By contrast, a putative DEVH box RNA helicase was identified as the first plant SG component in the course of characterizing an Arabidopsis plasmodesmata mutant [18]. Later on, it was shown that mammalian counterpart PB components (DCP1, DCP2, and XRN4) and SG components (eIF4E, RBP47b, and UBP1b) were localized in cytoplasmic mRNP granules in plants [19]. These results indicate that plants appear to contain at least two types of mRNP granules, but the distinction between PB and SG is still unclear. We have found that all 11 AtTZFs are localized in discrete cytoplasmic foci [20]. Since AtTZF1 co-localizes with putative PB and SG components, it raises a possibility that plant TZFs could localize to both PBs and SGs [9]. However, we have recently found that AtTZF1 does not co-localize with a plant PB component, raising a possibility that PB and SG compositions in plants are deviated from that of in mammals. Our current research is focused on the studies of composition, function, and assembly mechanism of plant P-bodies and stress granules.
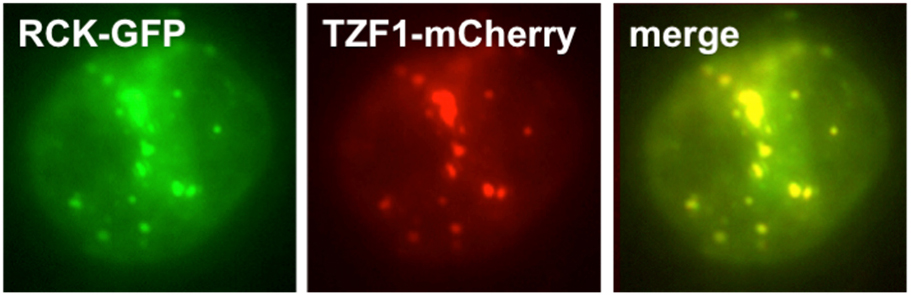
Figure 2. AtTZF1 co-localizes with a conserved mRNP granule marker RCK/Dhh1. Shown is a plant cell co-expressing two different fusion proteins, RCK-GFP and AtTZF1-mCherry. Merging of the two images shows complete co-localization of the two proteins in mRNP granules.
References
-
Jang, J.-C., and Sheen, J. (1997). Sugar sensing in higher plants. Trends in Plant Science 2, 208-214.
-
Sheen, J., Zhou, L., and Jang, J.C. (1999). Sugars as signaling molecules. Curr Opin Plant Biol 2, 410-418.
-
Jang, J.C., Leon, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 5-19.
-
Xiao, W., Sheen, J., and Jang, J.C. (2000). The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44, 451-461.
-
Zhou, L., Jang, J.C., Jones, T.L., and Sheen, J. (1998). Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci U S A 95, 10294-10299.
-
Price, J., Li, T.C., Kang, S.G., Na, J.K., and Jang, J.C. (2003). Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 132, 1424-1438.
-
Price, J., Laxmi, A., St Martin, S.K., and Jang, J.C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16, 2128-2150.
-
Kang, S.G., Price, J., Lin, P.C., Hong, J.C., and Jang, J.C. (2010). The Arabidopsis bZIP1 Transcription Factor Is Involved in Sugar Signaling, Protein Networking, and DNA Binding. Mol Plant 3, 361-373.
-
Pomeranz, M.C., Hah, C., Lin, P.C., Kang, S.G., Finer, J.J., Blackshear, P.J., and Jang, J.C. (2010). The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol 152, 151-165.
-
Lin, P.C., Pomeranz, M.C., Jikumaru, Y., Kang, S.G., Hah, C., Fujioka, S., Kamiya, Y., and Jang, J.C. (2011). The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J 65, 253-268.
-
Pomeranz, M., Zhang, L., Finer, J., and Jang, J.-C. (2011). Putative molecular mechanisms underlying tandem CCCH zinc finger proteins mediated plant growth, stress, and gene expression responses. Plant Signal Behav 6, 644-648.
-
Anderson, P., and Kedersha, N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10, 430-436.
-
Balagopal, V., and Parker, R. (2009). Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 21, 403-408.
-
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., and Anderson, P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169, 871-884.
-
Xu, J., Yang, J.Y., Niu, Q.W., and Chua, N.H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18, 3386-3398.
-
Goeres, D.C., Van Norman, J.M., Zhang, W., Fauver, N.A., Spencer, M.L., and Sieburth, L.E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19, 1549-1564.
-
Iwasaki, S., Takeda, A., Motose, H., and Watanabe, Y. (2007). Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett 581, 2455-2459.
-
Kobayashi, K., Otegui, M.S., Krishnakumar, S., Mindrinos, M., and Zambryski, P. (2007). INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19, 1885-1897.
-
Weber, C., Nover, L., and Fauth, M. (2008). Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J.
-
Pomeranz, M., Lin, P.C., Finer, J., and Jang, J.C. (2010). AtTZF gene family localizes to cytoplasmic foci. Plant Signal Behav 5, 190-192.
Jang Lab Members
-
Srimathi Bogamuwa (PhD graduate student)
-
Jie (Julie) Qu (PhD graduate student)
-
Thushani Rodrigo-Peiris (PhD graduate student)
