Susan Cole
Professor and Chair
282 Biological Sciences Building
484 West 12th Avenue
Columbus, OH
43210-1292
Areas of Expertise
- Vertebrate Development
- Cell Signaling
- Post-transcriptional Regulation
Research Interests
We are broadly interested in how cells communicate during embryonic development, how post-transcriptional regulation controls this communication, and how changes in cell signaling pathways influence congenital defects and diseases such as cancer. We have focused on the Notch signaling pathway; a highly conserved cell:cell communication pathway with multiple critical roles in embryonic development. Canonical Notch signaling appears elegantly simple: Notch receptors on a signal-receiving cell bind Notch ligands on a neighboring signal-sending cell, resulting in cleavage and release of the Notch intracellular domain (NICD). NICD translocates to the nucleus of the signal receiving cell and activates expression of Notch targets with no second messengers or signal amplification. Notch signaling is stoichiometric (one activated receptor creates one NICD molecule) and self-limiting (the signaling event destroys both the receptor and the ligand). Despite this apparent simplicity, Notch signaling is critical in a variety of developmental decisions, and it is clear that tight spatial and temporal control of the pathway is necessary in most of these contexts. We use techniques in genetics, molecular biology, mouse models, and embryology to address fundamental questions about how cell signaling is controlled during normal development and how changes in cell signaling influence human disease.
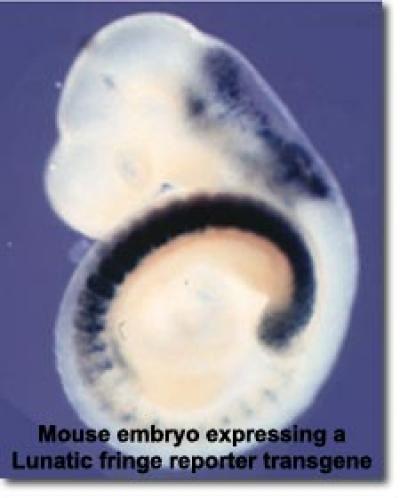
Oscillatory gene expression and Notch signaling: In several developmental contexts Notch pathway activity and Notch target expression exhibit rapid oscillations with periods ranging from two to six hours. These oscillations regulate the timing of key developmental outcomes, influencing embryonic patterning and cell fate decisions. For instance, the embryonic precursors of the vertebrae and ribs (somites) are produced by sequential budding from an overtly unsegmented tissue called the presomitic mesoderm (PSM). Coordinated oscillatory Notch activity in the PSM acts as part of a "segmentation clock" that times the production of somites. Cell autonomous oscillations have also been observed in neural stem cells during brain development and in embryonic stem cells. The regulation of oscillatory gene expression involves tight post-transcriptional regulation of Notch pathway components. Work from our lab and others has demonstrated that small changes in the expression or turnover of oscillatory gene products lead to significant phenotypic perturbations in the somites, ribs, and vertebrae. Previous and ongoing work from our lab has examined the transcriptional and post-transcriptional regulation of Lunatic fringe (a glycosyltransferase that modifies the Notch receptors and ligands), as well as the regulation of oscillatory Notch targets such as Hes1 and Hes7. Additional projects are defining the mechanisms by which Lunatic fringe and Deltalike-3 (an inhibitory Notch ligand) cooperate to regulate the segmentation clock.
Our findings and expertise have facilitated collaborations with labs at OSU and beyond examining the regulation of Notch signaling in a variety of contexts. With colleagues at Nationwide Children's we have examined how missense mutations in the Notch1 receptor may contribute to Left Ventricular Outflow Tract malformations in humans (McBride et al., 2008, Riley et al., 2011), and we have additional ongoing collaborations examining Notch regulation in cardiac development. Additional collaborations have examined the functions of MFNG during early endocardial development (D’Amato et al., 2015), and the roles of fringe family members in inner ear development (Basch et al. 2016) and intestinal homeostasis (Kadur Lakshminarasimha Murthy et al. 2018). Through long-standing connections to the OSU Comprehensive Cancer Center we have also contributed to studies examining Notch regulation in glioblastomas (Hu et al. 2012, Nandhu et al. 2014).
Laboratory Personnel:
Graduate Students: Lauren Levesque, Nathen Zavada, Yu Hsun (Wishing) Huang
Undergraduate Students: Lilly Wooden, Claire MacLaughlin
Publications:
- See a complete list of publications by Dr. Cole
- Bochter MS, Servello D, Kakuda S, D'Amico R, Ebetino MF, Haltiwanger RS, Cole SE. (2022) Lfng and Dll3 cooperate to modulate protein interactions in cis and coordinate oscillatory Notch pathway activation in the segmentation clock. Dev Biol. 487:42-56.
- Majumdar U, Manivannan S, Basu M, Ueyama Y, Blaser MC, Cameron E, McDermott MR, Lincoln J, Cole SE, Wood S, Aikawa E, Lilly B, Garg V. (2021) Nitric oxide prevents aortic valve calcification by S-nitrosylation of USP9X to activate NOTCH signaling. Sci Adv. Feb 5;7(6)
- Braunreiter K.M. and S.E.Cole. (2019) A tale of two clocks: phosphorylation of NICD by CDKs links cell cycle and segmentation clock. EMBO Rep. (invited commentary)
- Kadur Lakshminarasimha Murthy P, Srinivasan T, Bochter MS, Xi R, Varanko AK, Tung KL, Semerci F, Xu K, Maletic-Savatic M, Cole SE, and X. Shen (2018) Spatially Specific Fringe Modulation of Notch Ligands Supports Intestinal Homeostasis. Elife. Apr 9;7. pii: e35710
- Wahi, K., Friesen, S, Coppoa, V. and S.E. Cole. (2017) Putative binding sites for mir-125 family miRNAs in the mouse Lfng 3'UTR affect transcript expression in the segmentation clock, but mir-125a-5p is dispensable for normal somitogenesis. Developmental Dynamics. 246:740-748
- Basch M.L., Brown R.M., Jen H.I., Semerci F., Depreux F., Edlund R., Zhang H., Norton C.R., Gridley T., Cole S.E., Doetzlhofer A., Maletic-Savatic M., Segil N., and A.K. Groves. (2016) Fine-tuning of Notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates. Elife. Dec 14;5. pii: e19921
- Williams D.R., Shifley E.T., Braunreiter K.M., and S.E. Cole (2016). Disruption of somitogenesis by a novel dominant allele of Lfng suggests important roles for protein processing and secretion. Development 143:822-30.
- Wahi K., Bochter M.S., and S.E. Cole. (2016) The many roles of Notch signaling during vertebrate somitogenesis. Semin Cell Dev Biol. 49:68-75. (invited review)
- D'Amato G., Luxán G. Del Monte-Nieto G., Martínez-Poveda B., Torroja C. Walter W,. Bochter M.S., Benedito R., Cole S.E., Martinez F., Hadjantonakis A.K., Uemura A., Jiménez-Borreguero L.J., and J.L. de la Pompa. (2016) Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 18:7-20.
- Nandhu M.S., Hu, B., Cole S.E., Erdreich-Epstein, A., Rodriguez-Gil, D.J., and M. S. Viapiano. (2014) Novel paracrine modulation of Notch-DLL4 signaling by fibulin-3 promotes angiogenesis in high-grade gliomas. Cancer Res. 74:5435-48.
- Williams, D.R., Shifley, E.T., Lather, J.D. and S.E. Cole. (2014) Caudal skeletal development and the segmentation clock period are sensitive to Lfng dosage during somitogenesis. Dev. Biol. 388:159-69.
- Miller, A.J and S.E. Cole. (2014) Multiple Dlk1 splice variants are expressed during early mouse embryogenesis. Int. J. Dev. Biol. 58:65-70
- Riley, M.F., M.S. Bochter, K. Wahi, G.J. Nuovo and S.E. Cole. (2013) mir-125a-5p-mediated Regulation of Lfng is Essential for the Avian Segmentation Clock. Dev. Cell. 24:554-561 (Highlighted in Faculty of 1000)
- Hu, B., P. Agudelo-Garcia, H. Sim, J. Saldivar, C. Dolan, M. Mora, G. Nuovo, S.E. Cole, and M. Viapiano. (2012) Fibulin-3 promotes glioma growth and resistance through a novel paracrine regulation of Notch signaling. Cancer Res 72:3873-85.
- Riley, M.F., K.L. McBride and S.E. Cole. (2011) NOTCH1 missense alleles associated with left ventricular outflow tract defects exhibit impaired receptor processing and defective EMT. BBA Mol. Mech. Dis. 1812:121-9.
- Moran, J.L., E.T. Shifley, J.M. Levorse, S. Mani, K. Ostmann, A. Perez-Balaguer, D.M. Walker, T.F. Vogt, and S.E. Cole. (2009) Manic fringe is not required for embryonic development, and fringe family members do not exhibit redundant functions in the axial skeleton, limb, or hindbrain. Dev. Dyn. 238:1803-12.
- Shifley, E.T. and S.E. Cole. (2008) Lunatic fringe protein processing by proprotein convertases may contribute to the short protein half-life in the segmentation clock. Biochim Biophys Acta, Mol Cell Res. 1783, 2384-90.
- McBride, K.L., M.F. Riley, G.A. Zender, S.M. Fitzgerald-Butt, J.A. Towbin, J.W. Belmont and S.E. Cole. (2008) Notch1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol. Gen. 17, 2886-93.
- Ryan, M.J., C. Bales, A. Nelson, D.M. Gonzalez, L. Underkoffler, M. Segalov, J. Wilson-Rawls, S.E. Cole, J.L. Moran, P. Russo, N.B. Spinner, K. Kusumi, and K.M. Loomes. (2008) Bile Duct Proliferation in Jag1/Fringe Heterozygous Mice Identifies Candidate Modifiers of the Alagille Syndrome Hepatic Phenotype. Hepatology 48, 1989-1997.
- Shifley, E. T., K. M. Vanhorn, A. Perez-Balaguer, J. D. Franklin, M. Weinstein and S.E. Cole (2008) Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development 135, 899-908.
- Shifley, E. T. and S.E. Cole (2007) The vertebrate segmentation clock and its role in skeletal birth defects. Birth Defects Res C Embryo Today 81, 121-33. (invited review)
