Anna Dobritsa

Contact Information
Associate Professor
Areas of Expertise
- Pollen Development
- Cell Biology
- Plant Molecular Genetics
Cells rely on a regulated production of extracellular materials to control their shapes, growth, and motility, to promote tissue formation, and to protect themselves. Despite the importance of extracellular structures in development and disease, the question of how cells decide when, where, and in which manner these materials should be produced and deposited is far from being understood in any system. My laboratory studies formation of complex extracellular structures using the development of exine, the outer cell wall of plant pollen grains, as a model.
Exine, the amazing cell wall of pollen grains
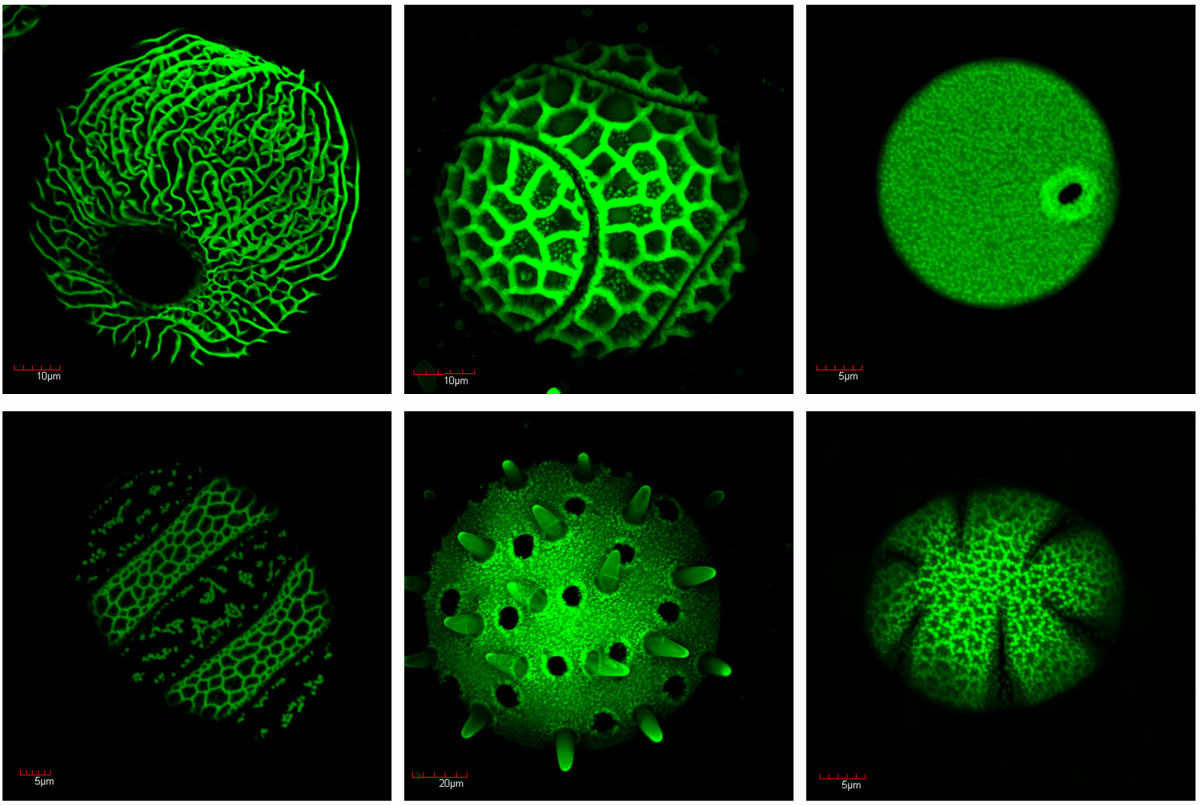
Figure. 1 Pollen exine assembles into elaborate species-specific patterns. Pollen grains of six plant species are shown.
Having evolved as a protector of male reproductive units, a pollinator-interacting entity, and a site of pollen-pistil recognition, exine is important for plant reproduction and speciation. Understanding the mechanism of its development could enable improved manipulations of plant reproduction by controlling pollination. In addition to this, exine formation is a fascinating system that lies at the intersection of many disciplines, ranging from cell and developmental biology to biochemistry, evolutionary biology, materials science, and molecular engineering. The following features make exine an exciting system to study:
- Exine has a very unique composition, different from other plant cell walls. It is made of sporopollenin, a biopolymer unparalleled for its strength, elasticity, and chemical durability. Sporopollenin’s chemical structure and the genetic and biochemical networks that control its production are poorly understood. Because of this biopolymer’s remarkable material properties, understanding the processes responsible for sporopollenin synthesis is of interest not only to plant biologists, but also to materials scientists.
- Unlike other plant cell walls that are directly produced by the cells that will be covered by these walls, exine formation depends on the unusual collaboration between the developing pollen and the other cell layers. This implies that there must be a lot of molecular dialog between the participating cells.
- Exine has an enormous morphological diversity across taxa and assembles into thousands of intricate species-specific 3D patterns (Fig. 1). How exine materials are assembled on pollen surfaces with such a precision into dramatically diverse patterns are the intriguing evo-devo and cell-biological questions.
Pollen apertures, a model for formation of distinct cellular domainsExine is typically deposited on the pollen surface non-uniformly, with the outcome that certain regions of the surface develop almost no exine. These areas (visible as darker regions in Fig.1) are known as pollen apertures – in many species, they are used as sites for pollen tube exit during germination. Similar to exine patterns, aperture patterns are generally conserved within a species but can vary considerably between the species, with apertures displaying different number, positions or morphology. Existence of apertures indicates that the pollen surface is not uniform and capable of creating distinct aperture domains that have a different molecular composition from the surrounding areas (Fig. 2). We are interested in molecular mechanisms involved in the formation of aperture domains and establishment of aperture patterns and have identified several genes involved in these processes.
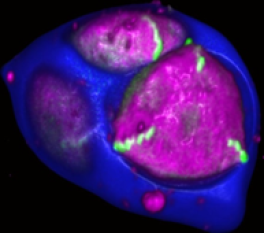
Figure 2. In developing pollen, the Arabidopsis INP1 protein (green) localizes specifically to three equally spaced regions of the plasma membrane, where it assembles into long and narrow lines. These regions develop into aperture domains that will become protected from exine deposition.
Research goals and approaches
We are trying to learn how sporopollenin is synthesized, exine is developed, exine and aperture patterns are laid out, and eventually would like to understand how the forms of these complex structures determine their functions. To answer these questions, we utilize genetics, molecular biology, confocal and electron microscopy, and biochemistry. We have a large collection of Arabidopsis exine and aperture mutants (some shown in Fig. 3) that help us to identify and study molecular players and events involved in sporopollenin biosynthesis and formation of exine and aperture patterns on the pollen surface. We have recently started using the data from our Arabidopsis research as a foundation for evo-devo studies regarding the generation of the enormous variety of exine patterns observed in nature.

Figure 3. Examples of phenotypes of our exine and aperture Arabidopsis mutants. Wild-type Arabidopsis pollen grains are in the top left panel. The inp1 mutant completely lacking apertures is the second in the top row.
LAB MEMBERS:
- Dr. Yuan Zhou
- Sarah Reeder, Research Associate
- Radhika Abeywickrama, undergraduate student, Molecular Genetics
- Laurel Fleszar, undergraduate student, Molecular Genetics
- Crystal Finzer, undergraduate student, Molecular Genetics

Dobritsa lab (Summer 2023): Sarah Reeder, Marco Keller, Crystal Finzer, Anna Dobritsa, Radhika Abeywickrama, Laurel Fleszar, and Yuan Zhou.
Dobritsa lab (Summer 2023): Yuan Zhou (and Ian), Anna Dobritsa, Crystal Finzer, Laurel Fleszar, Radhika Abeywickrama, and Sarah Reeder.
ALUMNI:
Postdoctoral Scholars
- Dr. Byungha Lee. 2014-2019
- Dr. Rui Wang, 2016-2021
Graduate students
- Galen Rask, 2015-2018
- Shayne Plourde (MS in Mathematical Biosciences), co-supervised with Adriana Dawes, 2016-2017
Visiting scholars
- Ingrid Moberg, Norwegian Unversity of Science and Technology, Ålesund, Norway, 2019
- Dr. Yunxia Chen, Forensic Center of Wildlife, Nanjing, China, 2017-2018
- Dr. Peng Li, Tsinghua University, China, 2015-2016
- Samira Ben-Menni Schuler, University of Granada, Spain, 2015
Undergraduate Assistants
- Marco Keller, NSF-REU student, University of Texas at Austin, 2023
- Mia Scott, OSU Biological Engineering, 2021 –2022
- Grace Hodson, OSU Molecular Genetics, 2021 –2022
- Jay Huang, OSU Biochemistry and Math, 2020 – May 2022
- Emili Toppari, Case Western Reserve University, 2021
- Stone Knapp, OSU Molecular Genetics, 2019- 2020
- Kern Lindsay, NSF REU student, 2019
- Thelma Amoah, OSU Biology, 2018-2019
- Jad Hussein, OSU Biomedical Engineering, 2017-2019
- Ayla Edwards, OSU Molecular Genetics, 2017-2018
- Claire Merriman, OSU Biology, 2016-2018
- Prativa Amom, OSU Molecular Genetics, 2015-2018
- Zachary Weber, OSU Molecular Genetics, 2014-2018
- Noah Weyrick, NSF/CAPS-REU student, 2018
- Michelle Tan, OSU Molecular Genetics, 2015-2017
- Holly Welfley, OSU Molecular Genetics, 2014-2016
- Ronnie Fox, OSU Molecular Genetics, 2014-2016
- Ruyuan Liu, OSU Biology, 2014-2015
- Matthew Postolowski, OSU Biology, 2014-2015
- Nahomie Chantal Rodriguez Sastre, NSF-REU student, 2014
High-school students
- Sydney Bernthold – K12, Metro School Internship, Metro High School, 2016
- Kevin Freeman – K12, Metro Capstone Program, Metro High School, 2014
Publications:

- Zhou, Y., Amom, P., Reeder, S. H., Lee, B. H., Helton, A., and A. A. Dobritsa (2021) Members of the ELMOD protein family specify formation of distinct aperture domains on the Arabidopsis pollen surface. eLife 10:e71061
- Wang, R., Owen, H. A., and A. A. Dobritsa (2021) Dynamic changes in primexine during the tetrad stage of pollen development. Plant Physiology 187: 2393-2404
- Lee, B. H., Wang, R., Moberg, I. M., Reeder, S. H., Amom, P., Tan, M. H., Amstutz, K., Chandna, P., Helton, A., Andrianova, E. P., Zhulin, I. B., and A. A. Dobritsa (2021) A species-specific functional module controls formation of pollen apertures. Nature Plants 7: 966-978
- Wang, R. and A. A. Dobritsa (2021) Loss of THIN EXINE2 disrupts multiple processes in the mechanism of pollen exine formation. Plant Physiology 187:133-157
- Zhou, Y. and A. A. Dobritsa (2020) Building portals in pollen. Nature Plants 6:334-335.
- Zhou, Y. and A. A. Dobritsa (2019) Formation of aperture sites on the pollen surface as a model for development of distinct cellular domains. Plant Science 288:110222
- Plourde, S., Amom, P., Tan, M., Dawes A. T., and A. A. Dobritsa (2019) Changes in morphogen kinetics and pollen grain size are potential mechanisms of aberrant pollen aperture patterning in previously observed and novel mutants of Arabidopsis thaliana. PLOS Computational Biology 15(2): e1006800.
- Lee, B. H., Weber, Z. T., Zourelidou, M., Hofmeister, B. T., Schmitz, R. J., Schwechheimer, C., and A. A. Dobritsa (2018) Arabidopsis protein kinase D6PKL3 is involved in the formation of distinct plasma-membrane aperture domains on the pollen surface. Plant Cell 30:2038-2056.
Wang, R. and A. A. Dobritsa (2018) Wang, R. and A. A. Dobritsa (2018) Exine and aperture patterns on the pollen surface: Their formation and roles in plant reproduction. Annual Plant Reviews 1: 1-40
- Dobritsa, A. A., Kirkpatrick, A., Reeder, S. H., Li, P. and H. A. Owen (2018) Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition. Plant Physiology. Focus Issue on Plant Cell Dynamics. 176: 326-339,
- Li, P., Ben-Menni Schuler, S., Reeder, S. H., Wang, R., Suarez Santiago, V., and A. A. Dobritsa (2018) INP1 involvement in pollen aperture formation is evolutionarily conserved and may require species-specific partners. J. Exp. Bot. 69: 983-996,
- Albert, B., Ressayre, A., Dillmann, C., Carlson, A., Swanson, R. J., Gouyon, P.-H., and A. A. Dobritsa (2018) Effect of aperture number on pollen germination, survival, and reproductive success in Arabidopsis thaliana. Ann. Bot. 121:733-740,
- Dobritsa, A. A. and S. H. Reeder (2017) Formation of pollen apertures in Arabidopsis requires an interplay between male meiosis, development of INP1-decorated plasma membrane domains, and the callose wall. Plant Signaling & Behavior. e1393136.
- Reeder, S. H., Lee, B. H., Fox, R., and A. A. Dobritsa (2016) A ploidy-sensitive mechanism regulates formation of apertures on the Arabidopsis pollen surface and guides localization of the aperture factor INP1. PLOS Genetics 12: e1006060
- Prieu, C., Matamoro-Vidal, A., Raquin, C., Dobritsa, A., Mercier, R., Gouyon, P.-H., and B. Albert (2016) Aperture number influences pollen survival in Arabidopsis mutants. Am. J. Bot. (Special Issue on the Ecology and Evolution of Pollen Performance) 103:432-439,
- Dobritsa, A. A. and D. Coerper (2012) The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine. Plant Cell 24: 4452-4464
- Dobritsa, A. A., Geanconteri, A., Shrestha, J., Carlson, A., Kooyers, N., Coerper, D., Urbanczyk-Wochniak, E., Bench, B. J., Sumner, L. W., Swanson, R., and D. Preuss (2011) A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiology 157: 947-970
- Dobritsa, A. A., Lei, Z., Nishikawa, S., Urbanczyk-Wochniak, E., Huhman, D. V., Preuss, D., and Sumner, L.W. (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen development in Arabidopsis thaliana. Plant Physiology 153: 937-955
- Dobritsa, A. A., Nishikawa, S., Preuss, D., Urbanczyk-Wochniak, E., Sumner, L. W., Hammond, A., Carlson, A.L., and Swanson, R. J. (2009) LAP3, a novel plant protein required for pollen development, is essential for pollen exine formation. Sex. Plant Reprod. 22: 167-177
- Dobritsa, A. A., Shrestha, J., Morant, M., Pinot, F., Matsuno, M., Swanson, R., Møller, B. L., and D. Preuss (2009) CYP704B1 is a long-chain fatty acid w-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis thaliana. Plant Physiology 151: 574-589
OUTREACH:
We have been participating in many outreach events at OSU and Columbus, such as Pollinator Palooza, COSI Science Festival, WestFest, State Science Day and others.

