Guramrit Singh
Contact Information
Associate Professor
Areas of Expertise
- Gene Expression
- RNA Processing
- Control of mRNA Fate
Control of post-transcriptional gene expression via dynamic ribonucleoprotein (RNP) complexes
Research Interests
Every cell relies on precise and efficient gene expression. Multiple layers of controls operate at transcriptional (DNA to RNA) and post-transcriptional (RNA to protein) levels to govern faithful expression of genes into proteins. Much of the post-transcriptional fate of an mRNA rests in the hands of proteins that complex with RNA to form ribonucleoproteins (mRNPs). mRNP assembly begins as early as the precursor-to-mRNA (pre-mRNA) is transcribed, and proceeds as pre-mRNA is sculpted into mRNA during several processing steps. mRNPs continue to evolve throughout their lifetime, shedding proteins and acquiring others as they move from one cellular compartment to another and/or as they are acted upon by numerous macromolecular machines (e.g. the nuclear pore, the translating ribosome). How these dynamic RNA-protein machineries assemble and function to control mRNA fate remains under intense investigation to fully understand the fidelity and accuracy of gene expression. Utilizing experimental tools ranging from cutting-edge RNA-Seq based methods to more traditional yet ever-powerful biochemical and molecular approaches, we are investigating the following post-transcriptional phenomena in cell culture and whole animal models:
-
Control of post-transcriptional gene expression by the Exon Junction Complex
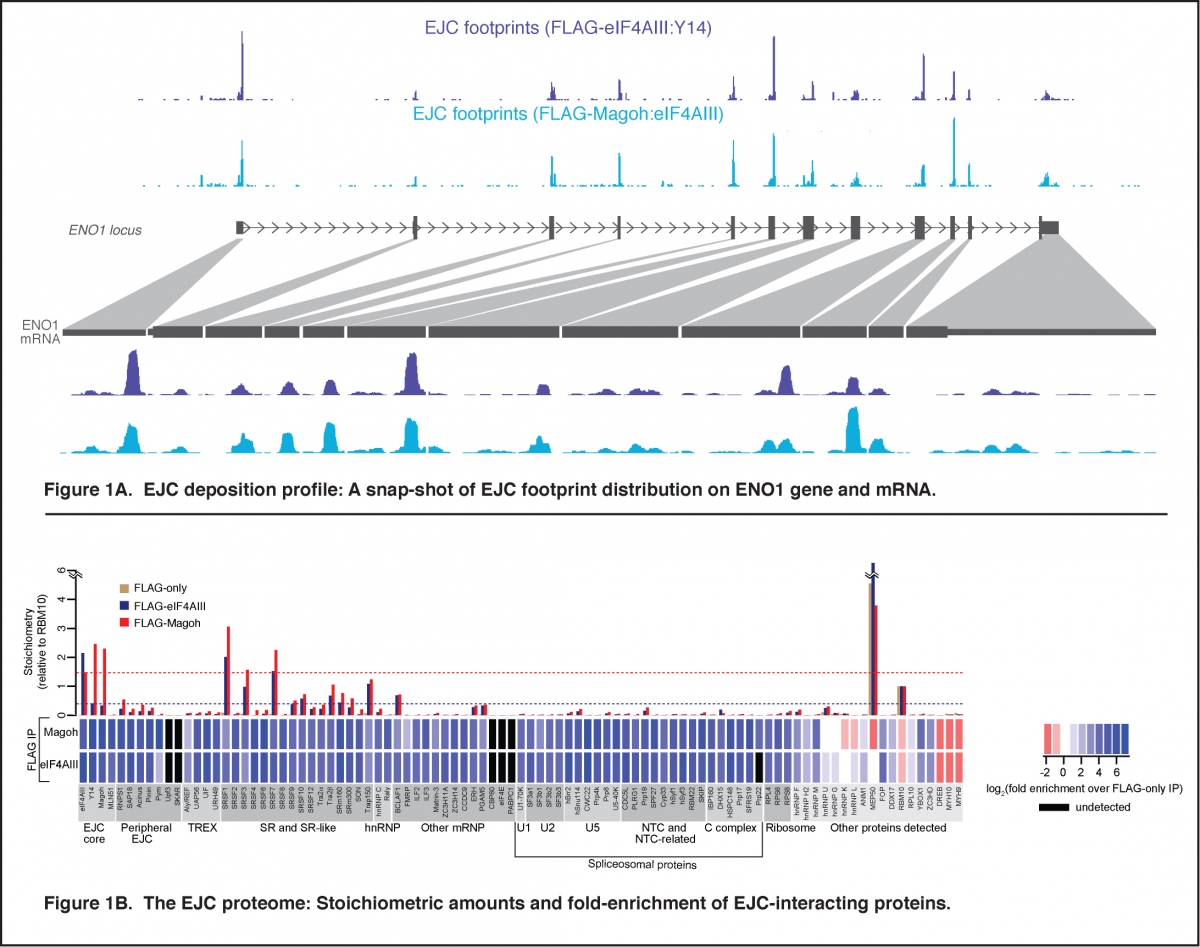
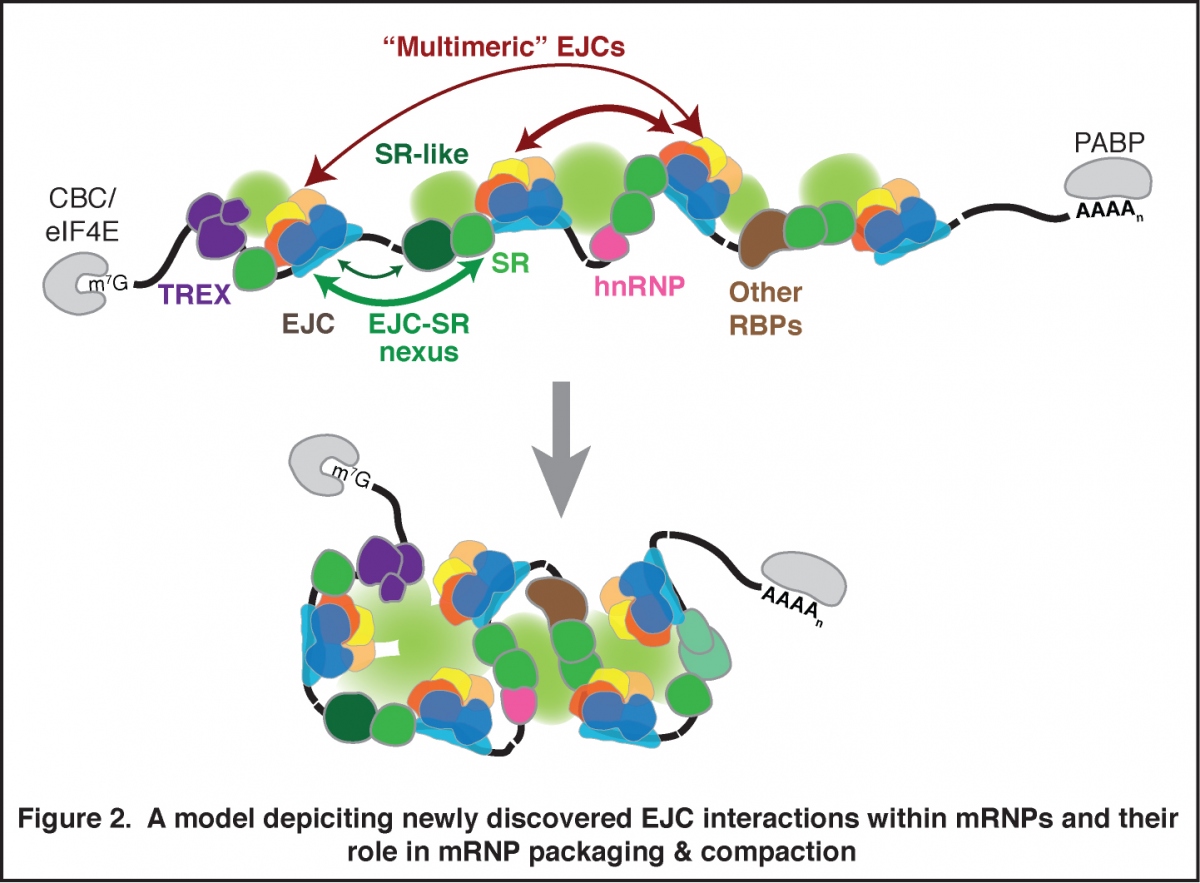
The Exon Junction Complex (EJC) is an extremely conserved multi-protein complex deposited ~24 nucleotides upstream of most mRNA exon-exon junctions during pre-mRNA splicing. The EJC is anchored on the RNA by eIF4AIII, which along with its co-factors, Y14, Magoh and MLN51, forms the EJC core. This core provides a platform for assembly of other peripheral EJC proteins that participate in pre-mRNA splicing and mRNA export from the nucleus, and in mRNA localization, translation and degradation in the cytoplasm. We previously uncovered the in vivo EJC interactome – its transcriptome-wide RNA binding sites and its proteome-wide interactions (Figure 1; Singh et al., Cell 2012). This work revealed that EJC is arguably the most prominent mRNP constituent as it occupies majority of exon junctions of human mRNAs. We have also discovered that in addition to the canonical exon junction sites, EJCs are also associated with non-canonical sites away from exon junctions. This connection is forged likely via EJC’s interactions with RNA binding proteins such as SR and SR-like proteins. We are now working to further understand composition of EJC at each assembly site at the transcriptome-wide scale, factors that influence EJC composition and how EJC composition impacts its function during post-transcriptional gene expression. We are also investigating the function of EJC and SR proteins in overall mRNP compaction, packaging and structure (Figure 2).
-
Developmental roles of the Exon Junction Complex in a vertebrate model (in collaboration with the Amacher lab)The extraordinarily conserved EJC proteins eIF4AIII, Y14, and Magoh play critical roles during animal development. Mutations in these proteins cause several human disorders: reduction in Y14 levels causes thrombocytopenia-absent radius syndrome, eIF4AIII mutation results in an autosomal recessive disorder with cleft mandible and limb anomalies, and copy number variation of EJC core proteins underlies X-linked mental retardation syndrome and intellectual disability. Additionally, Magoh haploinsufficiency in the mouse causes microcephaly and pigmentation defects. These observations hint at shared and non-overlapping roles for EJC core proteins and/or EJC core regulation by cell- or tissue-specific regulators. However, the precise role of the EJC in post-transcriptional gene regulation during development and differentiation is largely unknown. We are studying EJC core functions during embryonic development using zebrafish, a vertebrate model amenable to genetics, cell biology, and high throughput molecular biology and biochemistry. We are creating and characterizing zebrafish EJC null mutants to reveal whether these ubiquitously expressed proteins have broad or more tissue specific functions during development and/or if they play common or non-overlapping roles. Intriguingly, despite extreme conservation, the EJC is thought to function in NMD only in mammals; we are also testing whether the zebrafish EJC core is required for NMD. Alternative non-NMD functions of EJC that may regulate animal development are pre-mRNA splicing, mRNA export, localization, decay and translational regulation. Our RNA-Seq based molecular characterization of EJC core mutant embryos will assess some of these functions.
-
Translation initiation pathways in mammalsDuring early translation initiation steps, two eIF4A proteins that share high degree of identity with each other and with the EJC RNA clamp eIF4AIII, nucleate RNP machineries that unfurl the mRNA 5’-end to recruit the ribosome. eIF4AI protein is essential for cell survival whereas the role of eIF4AII protein remains largely unknown. Strikingly, eIF4AI and eIF4AII exhibit distinct expression patterns during mammalian development and during cellular response to internal or external stimuli. Contrary to the widely held belief, several lines of evidence point to an unequal role of the two proteins in translation initiation. Our objective is to uncover the cellular interactomes of eIF4AI and eIF4AII proteins to gain deeper insights into their function in translation, thus furthering our knowledge of translational control during cellular proliferation and transformation.
Lab Members

-
Tom Gallagher (Post-doc – joint with Dr. Sharon Amacher)
-
Lauren Woodward (Graduate Student)
-
Pooja Gangras (Graduate Student – co-mentored with Dr. Sharon Amacher)
-
Justin Mabin (Research Assistant)
Highly motivated students at all levels who are excited to study post-transcriptional gene regulation in mammalian cells are welcome to join our team. Several exciting projects are available in experimental areas at the interface of traditional molecular/cellular biology, biochemistry and computational biology. In addition to the exposure to multi-disciplinary research, we strive to provide a training environment that fosters independent thinking and creativity. If you wish to join our team, please contact Guramrit Singh.
Selected Publications
-
#Singh G., Pratt G., Yeo G.W., #Moore M.J. (2015) The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Ann. Rev. Biochem.,84:325-354 [#corresponding authors] [PMID: 25784054]
-
Ricci E.P., Kucukural A., Cenik C., Mercier B.C., Singh G., Heyer E.E., Ashar-Patel A., Peng L., Moore M.J. (2014) Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol., 21(1): 26-35 [PMID: 24336223]
-
Singh G., Ricci E. P. and Moore M. J. (2014) RIPiT-Seq: A high-throughput approach for footprinting RNA:protein complexes. Methods, 65(3): 320-332 [PMID: 24096052]
-
Kucukural A., Özadam H., Singh G., Moore M. J. and Cenik C. (2013) ASPeak: an abundance sensitive peak detection algorithm for RIP-Seq. Bioinformatics, doi: 10.1093/bioinformatics/btt428 [PMID: 23929032]
-
*Singh G., *Kucukural A., Cenik C., Leszyk, J. D., Shaffer, S. A., Weng Z. and Moore M. J. (2012) The cellular EJC interactome reveals higher order mRNP structure and an EJC-SR protein nexus. Cell, 151(4):750-764 [*equal contribution] [PMID: 23084401]
Highlighted in: Muhlemann O. (2012) Intimate liaison with SR proteins brings exon junction complexes to unexpected places. Nat Struct Mol Biol., 19(12):1209-1211 [PMID:23211765]
-
Morello L. G., Coltri P. P., Quaresma A. J., Simabuco F. M., Silva T. C., Singh G., Nickerson J. A., Oliveira, C. C., Moore M. J., and Zanchin, N. I. (2011) The human nucleolar protein FTSJ3 associates with NIP7 and functions in pre-rRNA processing. PLoS One, 2011;6(12):e29174 [PMID: 22195017]
-
Franks T. M., Singh G., and Lykke-Andersen J. (2010) Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell, 143(6): 938-950 [PMID: 21145460]
Highlighted in: Wilusz C. J. and Wilusz J. (2010) Consequences of mRNA wardrobe malfunctions. Cell, 143(6): 863-865 [PMID:21145451]
-
Singh G., Rebbapragada I., and Lykke-Andersen J. (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biology, 2008 Apr 29;6(4):e111 [PMID: 18447585]
-
Singh G., Jakob S., Kleedehn M., and Lykke-Andersen J. (2007) Communication with the exon junction complex and activation of nonsense mediated decay by the human Upf proteins occur in the cytoplasm. Molecular Cell, 27: 780-792 [PMID: 17803942]