David Somers
Contact Information
Professor
Areas of Expertise
- Plant Circadian Systems
- Post-transcriptional Mechanisms
- Phototransduction
Education
- Ph.D., University of California at Berkley, 1994.
The timing of many physiological and developmental processes in most eukaryotes is under the control of a circadian clock. This endogenous, self-sustaining oscillator maintains a rhythm of ca. 24 h in processes as diverse as human sleep/wake cycles, insect pupal eclosion, fungal sporulation and the movement of plant leaves. Many of the key events in plant development, such as flowering time, depend on receiving the appropriate environmental signals at the right time, and the circadian timekeeping mechanism allows them to keep pace with and anticipate cyclic events in their environment.
The central pacemaker driving circadian rhythms in plants consists of one or more autoregulatory feedback loops that are still being molecularly dissected. Work in my lab focuses on post-transcriptional control of the clock and in defining the molecular components that comprise the oscillator.
Post-translational regulation: proteostasis
ZEITLUPE (ZTL) is an F-box protein originally isolated as a long period mutant in Arabidopsis. It, and two related family members, are unique among the nearly 700 plant F-box proteins in possessing a blue-light sensing LOV (Light Oxygen and Voltage) domain at its N-terminus, very similar to the flavin-binding regions found in the phototropins. ZTL targets two members of a closely related family of pseudoresponse regulators (TOC1 and PRR5) that are key in setting the pace of the oscillator. Our work with ZTL has focused on understanding the role of each of three domains that comprise the protein, in the context of the circadian system. We identified ZTL as a novel blue light photoreceptor, the first F-box protein to possess this property (Kim et al., 2007).
Interactors with ZTL that regulates its stability in vivo are HSP90 and GIGANTEA (GI). GI has a wide-ranging role throughout plant development, including control over circadian period. GI confers post-translational control of circadian cycling to ZTL protein, acting as a co-chaperone with HSP90 to facilitate ZTL maturation. This finding identified a molecular role for GI and uncovered a new post-translational mechanism for establishing a circadian rhythm in eukaryotes (Cha et al., 2017).
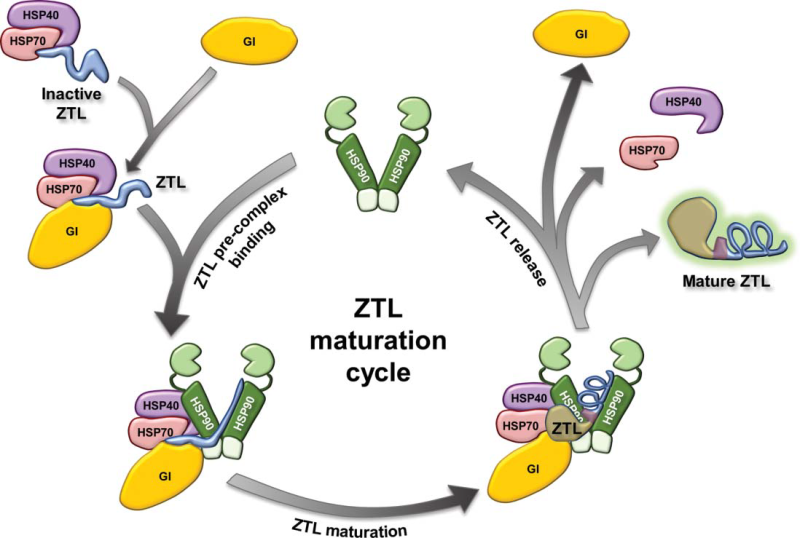
GIGANTEA as a co-chaperone essential to the maturation of the F-box protein ZEITLUPE
Model of the HSP90 chaperone cycle with the co-chaperone GIGANTEA acting on the client ZEITLUPE. Nascent ZTL is likely to first interact with HSP40/HSP70. GI is proposed to help introduce ZTL into the HSP90 chaperone cycle via interaction with the N-terminal LOV domain. Subsequently, an HSP90-GI-ZTL complex forms to facilitate maturation of ZTL. Not shown is the effect of blue light in facilitating the initial GI-ZTL interaction.
Post-translational regulation: phosphorylation
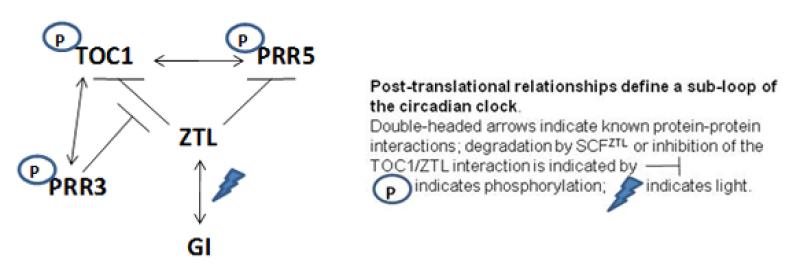
Role of protein phosphorylation in the control of period and robustness in the circadian clock. Five pseudoresponse regulator (PRR) proteins (TOC1, PRR3, PRR5, PRR7 and PRR9) are sequentially expressed over a 24 h diurnal and circadian cycle, and elimination and overexpression of each affects the period or robustness of the circadian oscillator. The phosphorylation state of most of them is also circadian regulated, and interaction between TOC1 and PRR3 depends on the phosphorylation state of both proteins (Fujiwara et al., 2008). The phosphorylation-based interaction between TOC1 and PRR3 competitively inhibits the TOC1/ZTL interaction, protecting TOC1 from SCFZTL-dependent degradation and altering circadian amplitude. In contrast, a TOC1/PRR5 interaction affects TOC1 phosphorylation, nuclear import and subnuclear localization (Wang et al., 2010). We are interested in understanding the role of the phosphorylation states of these proteins in regulating their interactions, and in their control of circadian period.
Teaching:
- Introductory Plant Physiology (MG 3436)
- Plant Physiology (MG 5630)
Lab Members:
Undergraduates
- Elizabeth Mouser
- Mackenzie Pritchard
Postdoctorates
- Dr. Breagha Magill
- Dr. Jingwen Yao
Lab Alumni:
- Dr. Jiapei Yan, National Key Laboratory of Crop Genetic Improvement Hubei, Huazhong Agricultural University, Wuhan China
- Dr. Hua Shi, State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan Agricultural University, Kunming, China
- Dr. Lei Wang, Assistant Professor, The Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing China
- Dr. Taesung Kim, Kongju National University, South Korea
- Dr. Lizhi Zhang, Postdoctoral Researcher, Department of Pharmacology, Ohio State University
- Dr. Woe-Yeon Kim, Assistant Professor, Gyeongsang National University, South Korea
- Dr. Jeongsik Kim, Group Leader, Daegu Gyeongbuk Institute of Science and Technology, South Korea
- Dr. Sumire Fujiwara, Group Leader, National Institute of Advanced Industrial Science and Technology, Japan
- Dr. Ruishuang Geng (Ph.D. 2006) Postdoctoral Researcher, Case Western Reserve University, Cleveland Ohio
- Dr. Linqu Han (Ph. D. 2006) Postdoctoral Researcher, University of Michigan
Selected Research Publications (2003 - Present):
- Yan J, Zhang Y, Chen G, Gui C, Tu Z, Cao D, Yao JW, Li X, Somers DE. TOC1 phosphorylation disproportionally enhances chromatin binding at rhythmic gene promoters. Sci Adv. 2025 Sep 26;11(39):eadx7804. doi: 10.1126/sciadv.adx7804.
- Hajdu A, Nyári DV, Ádám É, Kim YJ, Somers DE, Silhavy D, Nagy F, Kozma-Bognár L. Forward genetic approach identifies a phylogenetically conserved serine residue critical for the catalytic activity of UBIQUITIN-SPECIFIC PROTEASE 12 in Arabidopsis. Sci Rep. 2024 Oct 25;14(1):25273. doi: 10.1038/s41598-024-77232-w.
- Xie B, Luo M, Li Q, Shao J, Chen D, Somers DE, Tang D, Shi H. NUA positively regulates plant immunity by coordination with ESD4 to deSUMOylate TPR1 in Arabidopsis. New Phytol. 2024 Jan;241(1):363-377. doi: 10.1111/nph.19287.
- Somers DE. HSP90 in morphogenesis: taking the heat and keeping the dark. New Phytol. 2023 Aug;239(4):1157-1159. doi: 10.1111/nph.19062.
- Cha JY, Kim J, Jeong SY, Shin GI, Ji MG, Hwang JW, Khaleda L, Liao X, Ahn G, Park HJ, Kim DY, Pardo JM, Lee SY, Yun DJ, Somers DE, Kim WY. The Na(+)/H(+) antiporter SALT OVERLY SENSITIVE 1 regulates salt compensation of circadian rhythms by stabilizing GIGANTEA in Arabidopsis. Proc Natl Acad Sci U S A. 2022;119(33):e2207275119. Epub 2022/08/09. doi: 10.1073/pnas.2207275119.
- Pay ML, Kim DW, Somers DE, Kim JK, Foo M. Modelling of plant circadian clock for characterizing hypocotyl growth under different light quality conditions. In Silico Plants. 2022;4(1):diac001. Epub 2022/04/05. doi: 10.1093/insilicoplants/diac001
- Yan J, Li S, Kim YJ, Zeng Q, Radziejwoski A, Wang L, Nomura Y, Nakagami H, Somers DE. TOC1 clock protein phosphorylation controls complex formation with NF-YB/C to repress hypocotyl growth. EMBO J. 2021 Nov 2:e108684. doi: 10.15252/embj.2021108684
- Kim TS, Wang L, Kim YJ, Somers DE. Compensatory Mutations in GI and ZTL May Modulate Temperature Compensation in the Circadian Clock. Plant Physiol. 2020 Feb;182(2):1130-1141. doi: 10.1104/pp.19.01120.
- Kim YJ, Somers DE. Luciferase-Based Screen for Post-translational Control Factors in the Regulation of the Pseudo-Response Regulator PRR7. Front Plant Sci. 2019 May 22;10:667. doi: 10.3389/fpls.2019.00667
- Jo HH, Kim YJ, Kim JK, Foo M, Somers DE, Kim PJ. Waveforms of molecular oscillations reveal circadian timekeeping mechanisms. Commun Biol. 2018 Nov 26;1:207. doi: 10.1038/s42003-018-0217-1.
- Ritter A, Iñigo S, Fernández-Calvo P, Heyndrickx KS, Dhondt S, Shi H, De Milde L, Vanden Bossche R, De Clercq R, Eeckhout D, Ron M, Somers DE, Inzé D, Gevaert K, De Jaeger G, Vandepoele K, Pauwels L, Goossens A. The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat Commun. 2017 May 11;8:15235. doi: 10.1038/ncomms15235.
- Pudasaini A, Shim JS, Song YH, Shi H, Kiba T, Somers DE, Imaizumi T, Zoltowski BD. Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. Elife. 2017 Feb 28;6. pii: e21646. doi: 10.7554/eLife.21646.
- Cha JY, Kim J, Kim TS, Zeng Q, Wang L, Lee SY, Kim WY, Somers DE. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat Commun. 2017 Feb 23;8(1):3. doi: 10.1038/s41467-016-0014-9.
- Choudhary MK, Nomura Y, Shi H, Nakagami H, Somers DE. Circadian Profiling of the Arabidopsis Proteome Using 2D-DIGE. Front Plant Sci. 2016 Jul 12;7:1007. doi: 10.3389/fpls.2016.01007.
- Foo M, Somers DE, Kim PJ. 2016. Kernel architecture of the genetic circuitry of the Arabidopsis circadian system. PLoS.Comput.Biol. 12:e1004748.
- Choudhary MK, Nomura Y, Wang L, Nakagami H, Somers DE. 2015. Quantitative circadian phosphoproteomic analysis of Arabidopsis reveals extensive clock control of key components in physiological, metabolic and signaling pathways. Mol Cell Proteomics Aug;14(8):2243-60. doi: 10.1074/mcp.M114.047183.
- Kim J, Geng R, Gallenstein RA, Somers DE. 2013. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140(19):4060-9.
- Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM, Lin C. 2013. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci U S A. Oct 22;110(43):17582-7
- Kim Y, Han S, Yeom M, Kim H, Lim J, Cha JY, Kim WY, Somers DE, Putterill J, Nam HG, Hwang D. 2013. Balanced Nucleocytosolic Partitioning Defines a Spatial Network to Coordinate Circadian Physiology in Plants. Dev Cell. doi:pii: S1534-5807(13)00345-6. 10.1016/j.devcel.2013.06.006.
- Kim Y, Lim J, Yeom M, Kim H, Kim J, Wang L, Kim WY, Somers DE, Nam HG. 2013. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. Mar 28;3(3):671-7. doi: 10.1016/j.celrep.2013.02.021.
- Wang L, Kim J, Somers DE. 2013. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U. S. A 110: 761-766.
- Kim Y, Yeom M, Kim H, Lim J, Koo HJ, Hwang D, Somers D, Nam HG. 2012. GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol. Plant 5: 678-687.
- Kim TS, Kim WY, Fujiwara S, Kim J, Cha JY, Park JH, Lee SY, Somers DE. 2011. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc Natl Acad Sci U S A. Oct 4;108:16843-8. Epub 2011 Sep 26.
- Johansson M., McWatters H.G., Bakó L., Takata N., Gyula P., Hall A., Somers D.E., Millar A.J., Eriksson M.E. 2011. Partners in time: EARLY BIRD associates with ZEITLUPE and regulates the speed of the Arabidopsis clock. Plant Physiol. 155:2108-22.
- Kim, J and Somers D.E. 2010. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 154:611- 21.
- Wang , L. Fujiwara, S and Somers, D.E. 2010. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29:1903-15.
- Kim, W.Y., Salome, P.A., Fujiwara, S., Somers, D. E., and McClung, C.R. 2010. Characterization of pseudo- response regulators in plants. In Melvin, I.S. (ed.), Methods in Enzymology: Two-Component Signaling Systems, Part C, . Academic Press, pp. 357-378.
- Fujiwara, S., Wang, L., Han, L., Suh, S. S., Salome, P. A., McClung, C. R., Somers, D. E. 2008. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo- response regulator proteins. J.Biol.Chem. 283: 23073-23083
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM. 2008. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53(3):530-540
- Kim, W.Y, Fujiwara, S., Suh, S.S., Kim, J., Kim, Y., Han, L., David, K., Putterill, J., Nam. H.G., and Somers, D.E. 2007. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356-360.
- Allen T., Koustenis A., Theodorou G., Somers, D.E., Kay S.A., Whitelam G.C., Devlin P.F. 2006 Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18: 2506-2516[pdf]
- Kevei,E., Gyula,P., Hall,A., Kozma-Bognar,L., Kim,W.Y., Eriksson,M.E., Toth,R., Hanano,S., Feher,B., Southern,M.M., Bastow,R.M., Viczian,A., Hibberd,V., Davis,S.J., Somers, D.E., Nagy,F., and Millar,A.J. 2006 Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol 140:933-945 [pdf]
- Kim, W.Y., Hicks, K. A. and Somers, D.E. 2005. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 139:1557-69. [pdf]
- Han, L., Mason, M., Risseeuw, E.P., Crosby, W.L. and Somers, D.E. 2004. Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 40: 291-301. [pdf]
- Somers, D.E., Kim,W.Y., and Geng, R. 2004. The F-Box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769-782. [pdf]
- Mas P., Kim WY, Somers D.E., and Kay S.A. 2003. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 426:567-70. [pdf]
- Kim, W.Y., Geng, R., and Somers, D.E. 2003. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci U S A 100(8): 4933-4938. [pdf]
- Risseeuw, E.P., Daskalchuk, T.E., Banks, T.W., Liu, E., Cotelesage, J., Hellmann, H., Estelle, M., Somers, D.E., and Crosby, W.L. 2003. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34(6):753-767. [pdf]
Invited Publications, Book Chapters and Reviews (2005 - present)
- Yan, J., Kim, Y.J., Somers, D.E. Post-Translational Mechanisms of Plant Circadian Regulation. Genes (Basel) 2021, 12, doi:10.3390/genes12030325.
- Kim J, Somers DE. An HSP90 co-chaperone controls circadian proteostasis. Cell Cycle. 2017 Jul 19:1-2. doi: 10.1080/15384101.2017.1345238.
- Somers D.E. 2012. The Arabidopsis clock: time for an about-face? Genome Biol. 13: 153.
- Meier I, Somers D.E. 2011. Regulation of nucleocytoplasmic trafficking in plants.Curr Opin Plant Biol. 2011 Oct 14:538-46.
- Nelson, R.J. (ed.), Denlinger, D.L. (ed.), Somers, D. E. (ed.) 2010. Photoperiodism: The Biological Calendar. Oxford University Press, pp. 600.
- Somers, D. E. and Fujiwara, S. (2009) Thinking outside the F-box: novel ligands for novel receptors. Trends Plant Sci. 14(4):206-213.
- Somers, D. E., Fujiwara, S., Kim, W. Y., Suh, S. S. 2007. Post-translational photomodulation of circadian amplitude. Cold Spring Harb. Symp. Quant. Biol. 72: 193-200
- Somers, D.E. 2005. Entrainment of the Circadian Clock. In: Endogenous Plant Rhythms, eds. Hall, A.J.W and McWatters, H.G., Oxford: Blackwell, pp. 85-105
- Somers, D.E. 2005. ZEITLUPE and the Control of Circadian Timing. In: Light Sensing in Plants, eds.Wada, M., Shimazaki, K., and Iino, M., Tokyo: Springer, pp. 347-354..
[pdf] - Some links on this page are to .pdf files. These are designated by [pdf] following the link. pdf files require the use of Adobe Acrobat Reader software to open them. If you do not have Reader, you may use the following link to Adobe to download it for free.
