Desh Verma
Professor
240 Rightmire Hall
1060 Carmack Road
Columbus, OH
43210-1002
Areas of Expertise
- Plant Molecular Biology
- Stress Signaling
Education
- PhD, FRSC.
One of the research projects in my laboratory is aimed at understanding the mechanism involved in building the cell plate during cytokinesis in plant cells. The cell plate is built in the center of the phragmoplast, a microtubule held structure by fusion of Golgi-derived vesicles. While the homotypic fusion of small vesicles is likely to give rise to larger vesiclular structures, the vesicles at the cell plate fuse in a unique manner to give rise to a plate instead. This is apparently achieved by a recently identified protein in our laboratory, phragmoplastin, a homolog of dynamin. Phragmoplastin wraps around the vesicles to create dumbbell-shaped structures. A homotypic fusion of these structures is mediated by Knolle, a syntexin, and leads to the formation of a tubulovesicular network. This structure is filled by b-1,3 glucan (callose) which applies a sprea 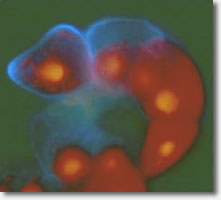 ding force that closes the holes in the tubular network and create a plate that grows from the center outwards. We have identified several partners of phragmoplastin which play roles in the organization and activation of the b-1-3 glucan synthase complex that begins de novo polysaccharide biosynthesis in this compartment. Since orientation and positioning of the cell plate is vital for plant organogenesis and architecture, particularly at the early stages of embryogenesis, understanding the steps involved in cell plate formation and the role of various players is essential. Finally, the existence of a family of phragmoplastin-like proteins suggests a wider role for this group of proteins in tubularization of membranes. Indeed, we have demonstrated that these proteins are targeted to different subcellular sites, suggesting a diverse role of this group of proteins. Using molecular, genetic and cellular fractionation approaches we plan to dissect the machinery involved in membrane fusion and in the triggering of callose biosynthesis during cell plate formation.
ding force that closes the holes in the tubular network and create a plate that grows from the center outwards. We have identified several partners of phragmoplastin which play roles in the organization and activation of the b-1-3 glucan synthase complex that begins de novo polysaccharide biosynthesis in this compartment. Since orientation and positioning of the cell plate is vital for plant organogenesis and architecture, particularly at the early stages of embryogenesis, understanding the steps involved in cell plate formation and the role of various players is essential. Finally, the existence of a family of phragmoplastin-like proteins suggests a wider role for this group of proteins in tubularization of membranes. Indeed, we have demonstrated that these proteins are targeted to different subcellular sites, suggesting a diverse role of this group of proteins. Using molecular, genetic and cellular fractionation approaches we plan to dissect the machinery involved in membrane fusion and in the triggering of callose biosynthesis during cell plate formation.
We are also studying the mechanism involved in osmotic stress regulation and osmoprotection in plants. The central question is how the stress signaling pathway is linked to the cell growth pathway. For this we have identified a plant TOR kinase that seems to act as a platform to integrate various stress signals and transduce them to respective effectors to control translation and transcription of specific genes involved in cell growth. This is a new area of study and holds significant potential for plant biology and biology at-large as this pathway controls cell growth in all eucaryotic organisms.

Verma Lab Members:
Post Doc: Dr. Zonglie Hong
PhD Students: Mr. Sunghon Kim, Mr. Magdy Mahfouz, Mr. Xiaoyun Dong
MSc Student: Mr. Chunbo Wang
Publications
- Kim Y-K, Kim S., Kim W-Y, Lee M-S, Cheon C-I and D. P. S. Verma(2012) Ribosomal Protein S6, a target of TOR kinase signaling, is involved in an epigenetic regulation of rRNA synthesis in Arabidopsis. (Submitted)
- Shin Y-J, Kim S., Du H.,Choi S., Verma DPS, Cheon-C-I (2012) Possible dual regulatory circuits involving AtS6K1 in the regulation of plant cell cycle and growth. Molecules and Cells 33: 487-496.
- Tan F, Zhang K, Mujahid H, Verma DPS and Peng Z. (2011) Differential histone modification and protein expression associated with cell wall removal and regeneration in rice (Oriza sativa). J. Proteome Research. 10: 551-563.
- Kim S., Kim S-J, Shin Y-, Kang J-H, Kim M-R, Hee N K, Lee M-S, Lee S-H, Kim Y-H, Hong S-K, Verma D. P. S., Chun J-Y, Cheon C-I (2009). An atypical soybean leucine-rich repeat receptor-like kinase, GmLRK1, may be involved in the regulation of cell elongation. Planta 229: 811-821.
- Dong Xiaoyun, Hong Zonglie, Chatterjee Jayanta, Kim Sunghan, Verma Desh Pal S (2008) Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta, vol.229 pp:87-98
- Lian Ma, Bo Xie, Zonglie Hong, Desh Pal S. Verma, and Zhongming Zhang (2008), A Novel RNA-Binding Protein Associatedwith Cell Plate Formation, Plant Physiology,Vol. 148, pp. 223–234.
- Dong X, and D. P. S. Verma (2008) Tissue-specific expression of Callose synthase genes. Planta 229, 87-98.
- Mahfouz M., Kim S, Delauney A J. and Verma D P S. (2006) Arabidopsis TARGET OF RAPAMYCIN Interacts with RAPTOR, Which Regulates the Activity of S6 Kinase in Response to Osmotic Stress Signal. Plant Cell 18:477-490
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315-328.
- Geisler-Lee J, Hong Z, Verma DPS (2002) Overexpression of phragmoplastin results in increased level of callose deposition at he forming cell plate. Plant Science 163: 33-42
- Hong, Z., A.J. Delauney, and D.P.S. Verma. (2001) A cell plate-specific callose synthase and its interaction with phragmoplastin and UDP-glucose transferase. Plant Cell 13, 755-768.
- Hong, Z, J. Olson, and D. P. S. Verma. (2001) A novel UDP glucose transferase interacts with callose synthase and phragmoplastin at the forming cell plate. Plant Cell 13, 769-780.
- Verma, D.P.S. (2001) Cytokinesis and Building of the Cell Plate in Plants. Annual Rev. of Plant Physiol. and Plant Molecular Biology 52: 751-784.
- Zhang, Z., Z. Hong, and D.P.S. Verma (2000) Phragmoplastin polymerizes in to spiral coiled structures via two self assembly domins J. Biol. Chem. 275: 8779-8784.
